Code
library(tidyverse)
library(easystats)
library(patchwork)
library(ggside)library(tidyverse)
library(easystats)
library(patchwork)
library(ggside)df <- read.csv("../data/data_raw.csv") |>
mutate(across(everything(), ~ifelse(.x == "", NA, .x))) |>
filter(Age >= 18) |>
mutate(Age = ifelse(Age > 90, NA, Age))The initial sample consisted of 1053 participants (Mean age = 24.1, SD = 10.5, range: [18, 76], 9.5e-02% missing; Gender: 75.6% women, 21.0% men, 2.56% non-binary, 0.85% missing; Education: Bachelor, 22.41%; Doctorate, 1.90%; High School, 65.53%; Master, 5.22%; missing, 0.28%; Other, 4.65%).
# Consecutive count of participants per day (as area)
df |>
mutate(Date = as.Date(Date, format = "%d/%m/%Y")) |>
group_by(Date, Experimenter) |>
summarize(N = n()) |>
ungroup() |>
complete(Date, Experimenter, fill = list(N = 0)) |>
group_by(Experimenter) |>
mutate(N = cumsum(N)) |>
ggplot(aes(x = Date, y = N)) +
geom_area(aes(fill=Experimenter)) +
scale_y_continuous(expand = c(0, 0)) +
labs(
title = "Recruitment History",
x = "Date",
y = "Total Number of Participants"
) +
see::theme_modern()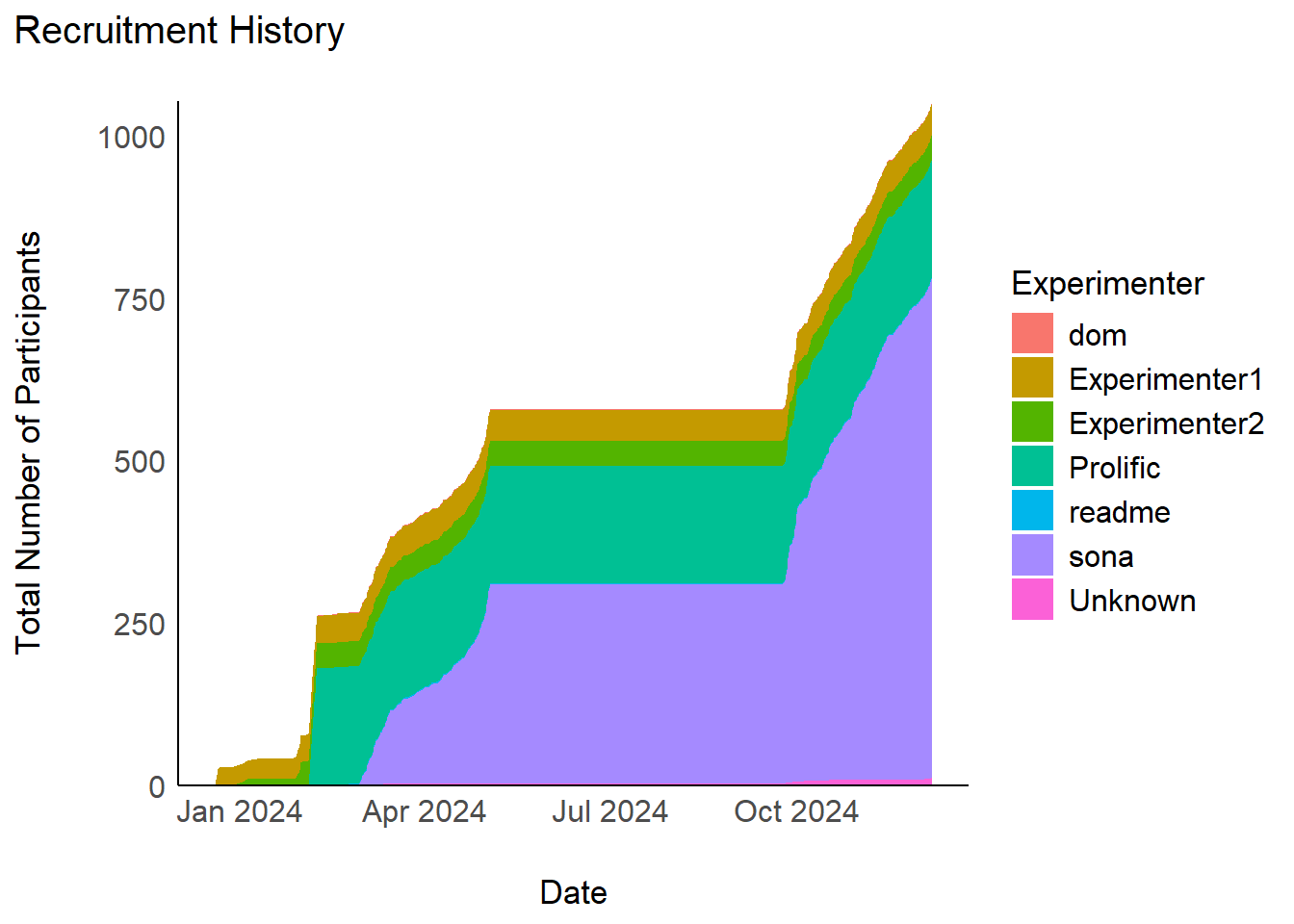
# Table
summarize(df, N = n(), .by=c("Experimenter")) |>
arrange(desc(N)) |>
gt::gt() |>
gt::opt_stylize() |>
gt::opt_interactive(use_compact_mode = TRUE) |>
gt::tab_header("Number of participants per recruitment source")df$Sample <- case_when(
df$Experimenter %in% c("Experimenter1", "Experimenter2") ~ "Student",
df$Experimenter %in% c("dom", "Unknown", "readme") ~ "Other",
.default = df$Experimenter
)The experiment’s median duration is 9.95 min (50% HDI [5.61, 11.80]).
df |>
mutate(Participant = fct_reorder(Participant, Experiment_Duration),
Category = ifelse(Experiment_Duration > 30, "extra", "ok"),
Duration = ifelse(Experiment_Duration > 30, 30, Experiment_Duration)) |>
ggplot(aes(y = Participant, x = Duration)) +
geom_vline(xintercept = median(df$Experiment_Duration), color = "red", linetype = "dashed") +
geom_point(aes(color = Sample, shape = Category)) +
geom_text(aes(color = Sample, label=ifelse(Experiment_Duration < 3, as.character(Participant), '')), hjust=-1, vjust=0) +
scale_shape_manual(values = c("extra" = 3, ok = 19)) +
guides(shape = "none") +
ggside::geom_xsidedensity(aes(fill=Sample), color=NA, alpha=0.5) +
ggside::scale_xsidey_continuous(expand = c(0, 0)) +
labs(
title = "Experiment Completion Time",
x = "Duration (in minutes)",
y = "Participant"
) +
theme_bw() +
ggside::theme_ggside_void() +
theme(ggside.panel.scale = .3,
panel.border = element_blank(),
axis.ticks.y = element_blank(),
axis.text.y = element_blank()) 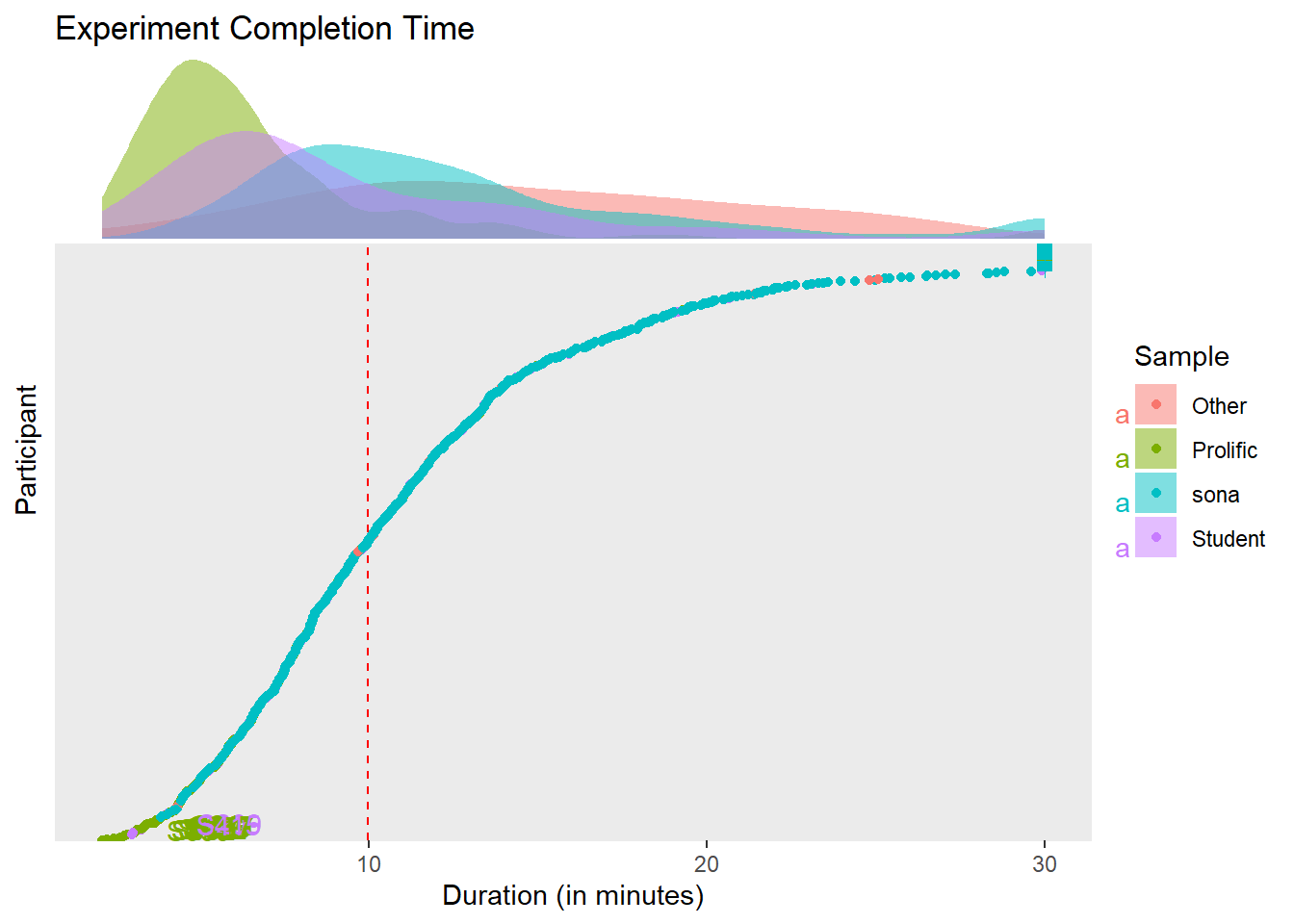
recode_phq <- function(x) {
case_when(
x == 1 ~ 0.5,
x == 2 ~ 1,
x == 3 ~ 2,
x == 4 ~ 3,
.default = x
)
}
df <- df |>
mutate(
PHQ4_Anxiety_1 = ifelse(PHQ4_Condition == "PHQ4 - Revised", recode_phq(PHQ4_Anxiety_1), PHQ4_Anxiety_1),
PHQ4_Anxiety_2 = ifelse(PHQ4_Condition == "PHQ4 - Revised", recode_phq(PHQ4_Anxiety_2), PHQ4_Anxiety_2),
PHQ4_Depression_3 = ifelse(PHQ4_Condition == "PHQ4 - Revised", recode_phq(PHQ4_Depression_3), PHQ4_Depression_3),
PHQ4_Depression_4 = ifelse(PHQ4_Condition == "PHQ4 - Revised", recode_phq(PHQ4_Depression_4), PHQ4_Depression_4),
PHQ4_Anxiety = PHQ4_Anxiety_1 + PHQ4_Anxiety_2,
PHQ4_Depression = PHQ4_Depression_3 + PHQ4_Depression_4,
PHQ4_Total = PHQ4_Anxiety + PHQ4_Depression
)
df <- df |>
mutate(
Weighted_PHQ4_Anxiety_1 = ifelse(
PHQ4_Condition == "PHQ4 - Revised",
case_when(
PHQ4_Anxiety_1 == 0 ~ 0,
PHQ4_Anxiety_1 == 0.5 ~ 0.34,
PHQ4_Anxiety_1 == 1 ~ 0.55,
PHQ4_Anxiety_1 == 2 ~ 0.71,
PHQ4_Anxiety_1 == 3 ~ 1,
.default = PHQ4_Anxiety_1
), PHQ4_Anxiety_1
),
Weighted_PHQ4_Anxiety_2 = ifelse(
PHQ4_Condition == "PHQ4 - Revised",
case_when(
PHQ4_Anxiety_2 == 0 ~ 0,
PHQ4_Anxiety_2 == 0.5 ~ 0.44,
PHQ4_Anxiety_2 == 1 ~ 0.58,
PHQ4_Anxiety_2 == 2 ~ 0.71,
PHQ4_Anxiety_2 == 3 ~ 1,
.default = PHQ4_Anxiety_2
), PHQ4_Anxiety_2
),
Weighted_PHQ4_Depression_3 = ifelse(
PHQ4_Condition == "PHQ4 - Revised",
case_when(
PHQ4_Depression_3 == 0 ~ 0,
PHQ4_Depression_3 == 0.5 ~ 0.38,
PHQ4_Depression_3 == 1 ~ 0.51,
PHQ4_Depression_3 == 2 ~ 0.62,
PHQ4_Depression_3 == 3 ~ 1,
.default = PHQ4_Depression_3
), PHQ4_Depression_3
),
Weighted_PHQ4_Depression_4 = ifelse(
PHQ4_Condition == "PHQ4 - Revised",
case_when(
PHQ4_Depression_4 == 0 ~ 0,
PHQ4_Depression_4 == 0.5 ~ 0.35,
PHQ4_Depression_4 == 1 ~ 0.52,
PHQ4_Depression_4 == 2 ~ 0.66,
PHQ4_Depression_4 == 3 ~ 1,
.default = PHQ4_Depression_4
), PHQ4_Depression_4
),
Weighted_PHQ4_Anxiety = ifelse(
PHQ4_Condition == "PHQ4 - Revised", datawizard::rescale(Weighted_PHQ4_Anxiety_1 + Weighted_PHQ4_Anxiety_2, range = c(0, 2), to = c(0, 6)), PHQ4_Anxiety_1 + PHQ4_Anxiety_2
),
Weighted_PHQ4_Depression = ifelse(
PHQ4_Condition == "PHQ4 - Revised", datawizard::rescale(Weighted_PHQ4_Depression_3 + Weighted_PHQ4_Depression_4, range = c(0, 2), to = c(0, 6)), PHQ4_Depression_3 + PHQ4_Depression_4
),
Weighted_PHQ4_Total = Weighted_PHQ4_Anxiety + Weighted_PHQ4_Depression
)
# df |>
# select(starts_with("Weighted_PHQ4_"), PHQ4_Condition) |>
# pivot_longer(cols = -PHQ4_Condition, names_to = "item", values_to = "score") |>
# ggplot(aes(x = score)) +
# geom_density(fill="#9C27B0", adjust=1/2) +
# facet_grid(item ~ PHQ4_Condition)df |>
select(starts_with("PHQ4_")) |>
pivot_longer(cols = matches("Depression_|Anxiety_"), names_to = "item", values_to = "score") |>
ggplot(aes(x = as.factor(score))) +
geom_bar(stat="count", fill="#9C27B0") +
facet_grid(item ~ PHQ4_Condition) +
labs(x = "Score", y = "N answers") +
theme_bw()
df |>
select(matches("STAI5_"), -STAI5_Duration, -STAI5_Order) |>
pivot_longer(cols = matches("STAI5_"), names_to = "item", values_to = "score") |>
ggplot(aes(x = as.factor(score))) +
geom_bar(stat="count", fill="#E91E63") +
facet_wrap(~item) +
labs(x = "Score", y = "N answers") +
theme_bw()
df <- df |>
mutate(STAI5_General = (STAI5_1 + STAI5_2 + STAI5_3 + STAI5_4 + STAI5_5) / 5)df |>
select(matches("BDI2_"), -BDI2_Duration, -BDI2_Order) |>
pivot_longer(cols = matches("BDI2_"), names_to = "item", values_to = "score") |>
ggplot(aes(x = as.factor(score))) +
geom_bar(stat="count", fill="#2196F3") +
facet_wrap(~item) +
labs(x = "Score", y = "N answers") +
theme_bw()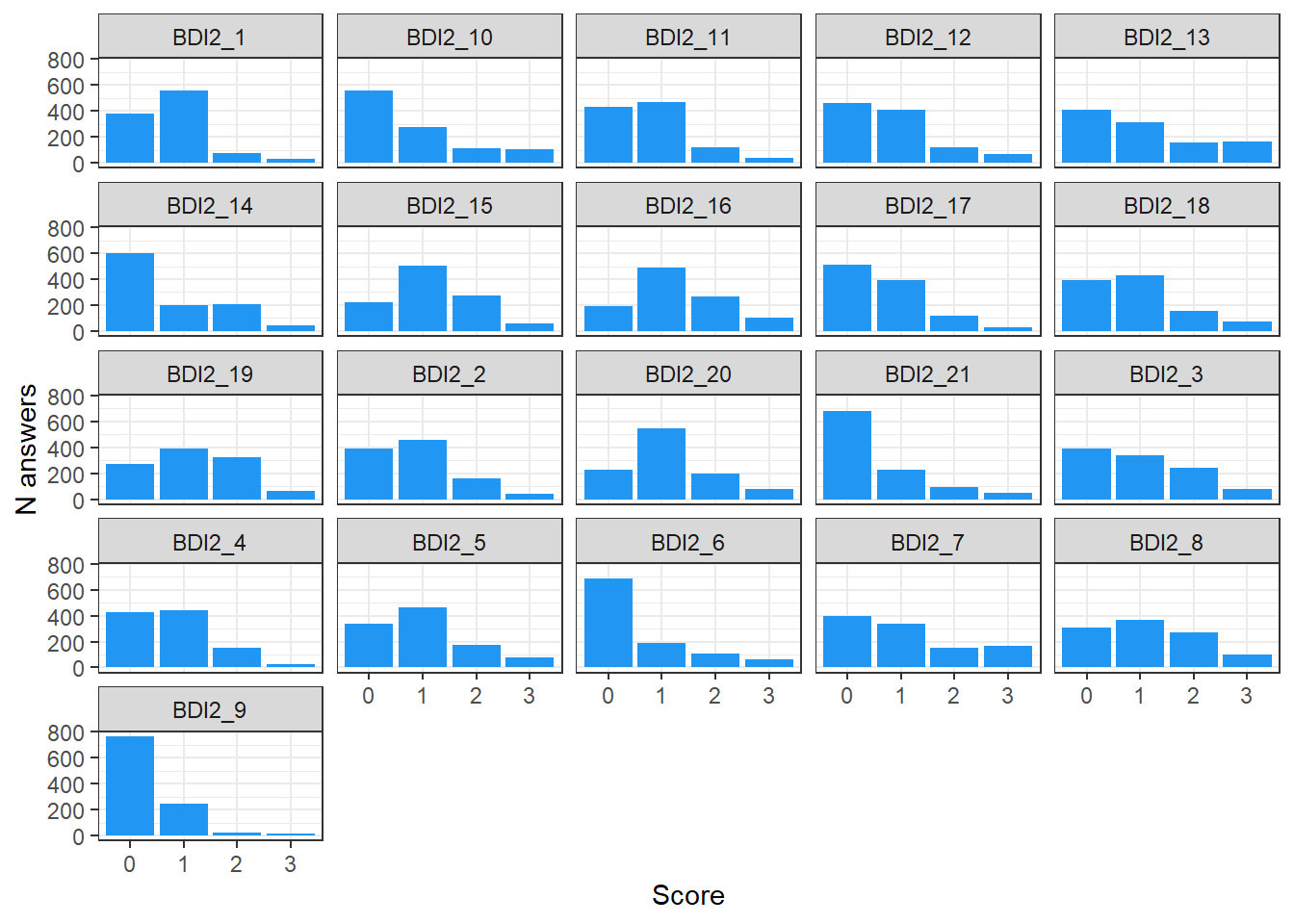
df <- df |>
mutate(BDI2_Total = rowSums(select(df, matches("BDI2_"), -BDI2_Duration, -BDI2_Order)))df <- df |>
mutate(IAS_Total = rowSums(select(df, matches("IAS_"), -IAS_Duration, -IAS_Order)))outliers <- list()Only a subset of participants saw the version with attention checks.
reject <- df |>
filter(!is.na(AttentionCheck_2)) |>
select(Participant, Experimenter, starts_with("AttentionCheck")) |>
arrange(AttentionCheck_2, AttentionCheck_3)
outliers$attentionchecks <- reject[reject$AttentionCheck_2 == 0 | reject$AttentionCheck_3 < 0.95, "Participant"]
outliers$attentionchecks <- outliers$attentionchecks[!is.na(outliers$attentionchecks)]
df$AttentionCheck <- ifelse(df$Participant %in% outliers$attentionchecks, "Failed", "Passed")
reject |>
gt::gt() |>
gt::data_color(columns = c("AttentionCheck_2", "AttentionCheck_3"), palette=c("red", "green")) |>
gt::data_color(columns = "Participant", fn=\(x) ifelse(x %in% outliers$attentionchecks, "red", "white")) |>
gt::opt_interactive(use_compact_mode = TRUE)We removed 194 (18.42%) participants for failing the attention checks.
dat <- mutate(df, Failed = ifelse(Participant %in% outliers$attentionchecks, 1, 0)) |>
filter(Gender %in% c("Female", "Male")) |>
filter(Experiment_Duration < 30) |>
mutate(Gender = as.factor(Gender)) |>
select(Failed, Gender, Experiment_Duration, Age, Ethnicity)
m <- glm(Failed ~ Gender / Age, data=dat, family="binomial")
parameters::parameters(m)
modelbased::estimate_relation(m, length=50) |>
ggplot(aes(x=Age, y=Predicted, color=Gender)) +
geom_line() m <- mgcv::gam(Failed ~ s(Experiment_Duration),
data=df |>
filter(Experiment_Duration < 30) |>
mutate(Failed = ifelse(AttentionCheck == "Failed", 1, 0)),
family="binomial")
parameters::parameters(m)# Fixed Effects
Parameter | Log-Odds | SE | 95% CI | z | df | p
--------------------------------------------------------------------------
(Intercept) | -1.52 | 0.08 | [-1.68, -1.36] | -18.46 | 1004.00 | < .001
# Smooth Terms
Parameter | z | df | p
-------------------------------------------------------
Smooth term (Experiment Duration) | 2.84 | 1.00 | 0.092
The model has a log- or logit-link. Consider using `exponentiate =
TRUE` to interpret coefficients as ratios.plot(estimate_relation(m, length=40))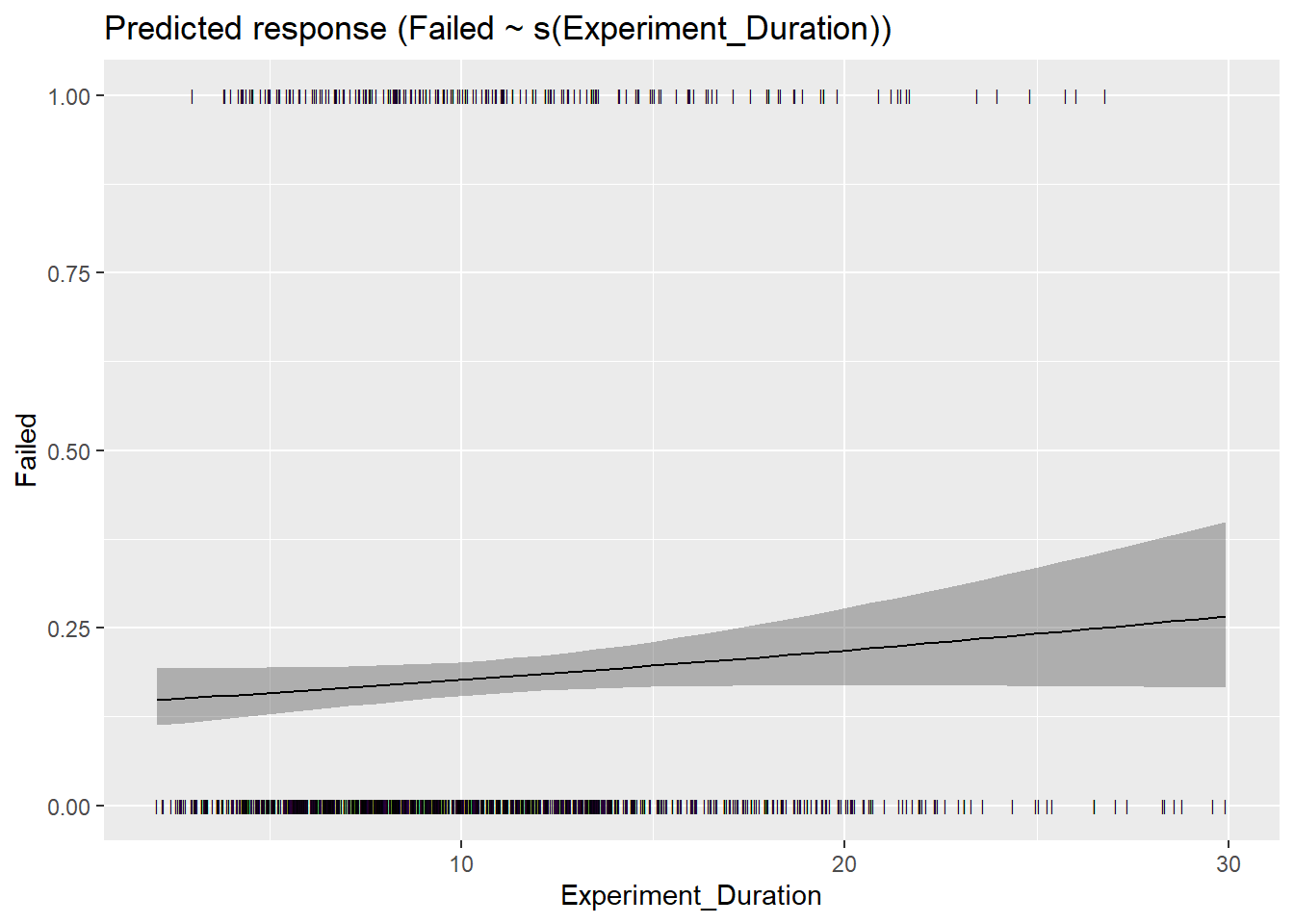
df |>
filter(Experiment_Duration < 30) |>
select(Participant, Experiment_Duration, AttentionCheck) |>
mutate(Participant = fct_reorder(Participant, Experiment_Duration)) |>
ggplot(aes(x=Participant, y=Experiment_Duration, fill=AttentionCheck)) +
geom_bar(stat="identity") +
theme_minimal() +
theme(axis.text.x = element_blank())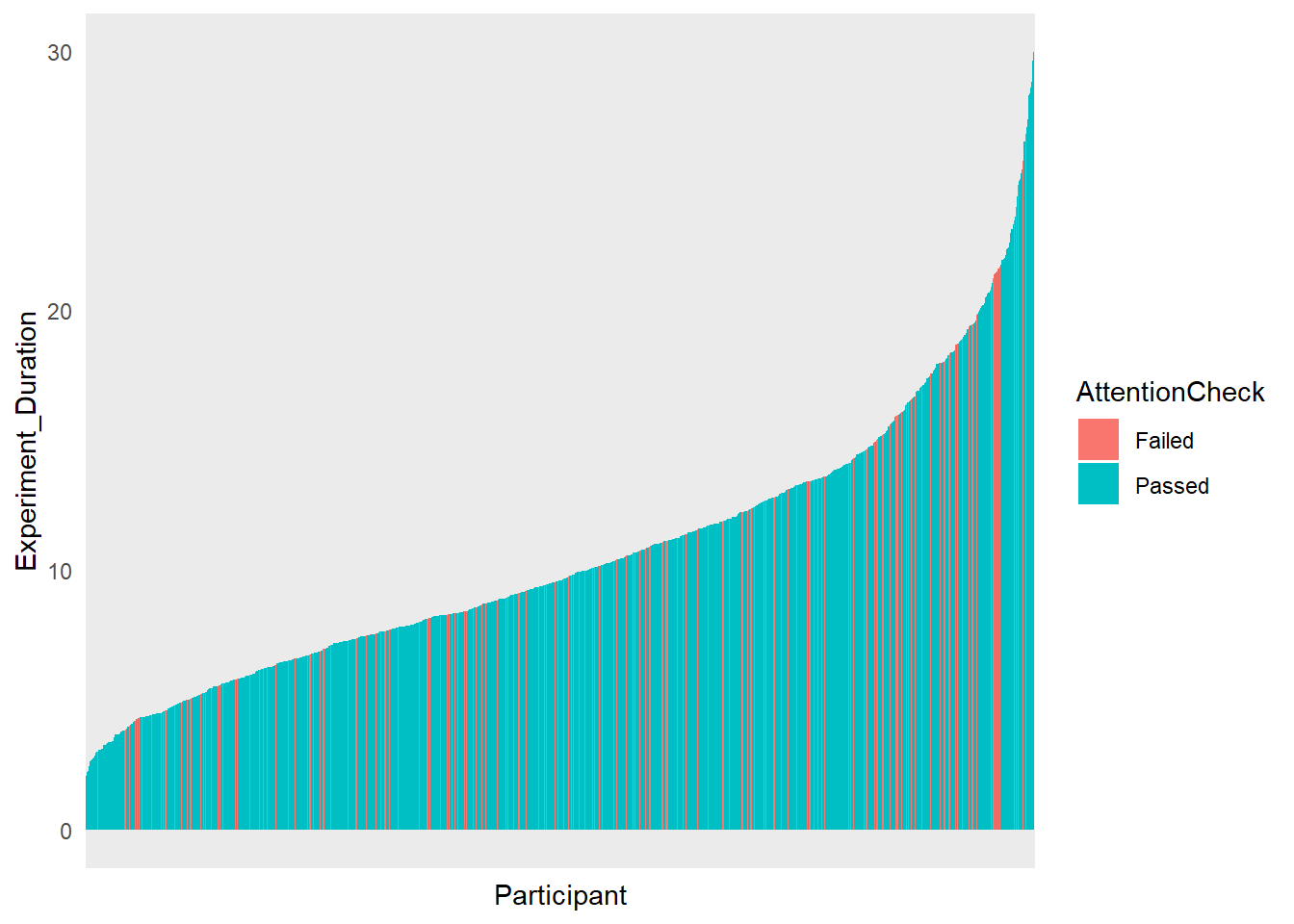
SD_per_dim <- function(x, dims="") {
m <- matrix(nrow=nrow(x), ncol=0)
for(s in dims) {
m <- cbind(m, sapply(as.data.frame(t(x[grepl(s, names(x))])), sd))
}
m
}
df$BDI2_SD <- select(df, matches("BDI2_"), -BDI2_Duration, -BDI2_Order, -BDI2_Total) |>
SD_per_dim(dims=c("BDI2_")) |>
rowMeans()
df$STAI5_SD <- select(df, matches("STAI5_"), -STAI5_Duration, -STAI5_Order, -STAI5_General) |>
SD_per_dim(dims=c("STAI5_")) |>
rowMeans()
df$IAS_SD <- select(df, matches("IAS_"), -IAS_Duration, -IAS_Order, -IAS_Total) |>
SD_per_dim(dims=c("IAS_")) |>
rowMeans()m <- mgcv::gam(Failed ~ s(IAS_SD),
data=mutate(df, Failed = ifelse(AttentionCheck == "Failed", 1, 0)),
family="binomial")
parameters::parameters(m)# Fixed Effects
Parameter | Log-Odds | SE | 95% CI | z | df | p
--------------------------------------------------------------------------
(Intercept) | -1.51 | 0.08 | [-1.67, -1.35] | -18.63 | 1051.00 | < .001
# Smooth Terms
Parameter | z | df | p
--------------------------------------------
Smooth term (IAS SD) | 11.84 | 1.00 | < .001estimate_relation(m, length=40) |>
ggplot(aes(x=IAS_SD, y=Predicted)) +
geom_ribbon(aes(ymin=CI_low, ymax=CI_high), alpha=0.2) +
geom_line() +
ggside::geom_xsidedensity(data=df, aes(fill=AttentionCheck), alpha=0.3, color=NA)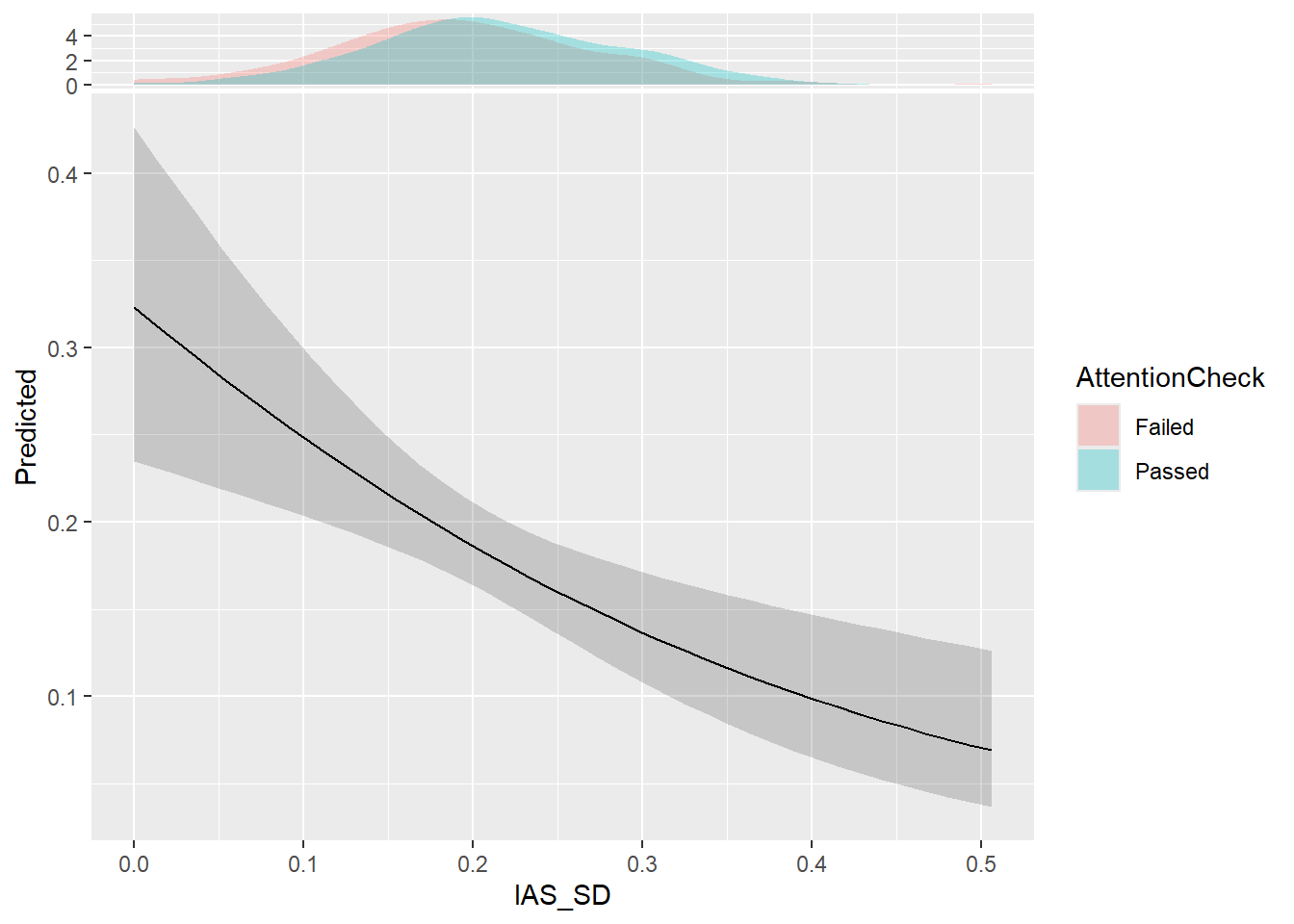
df |>
select(Participant, IAS_SD, AttentionCheck) |>
mutate(Participant = fct_reorder(Participant, IAS_SD)) |>
ggplot(aes(x=Participant, y=IAS_SD, fill=AttentionCheck)) +
geom_bar(stat="identity") +
theme_minimal() +
theme(axis.text.x = element_blank())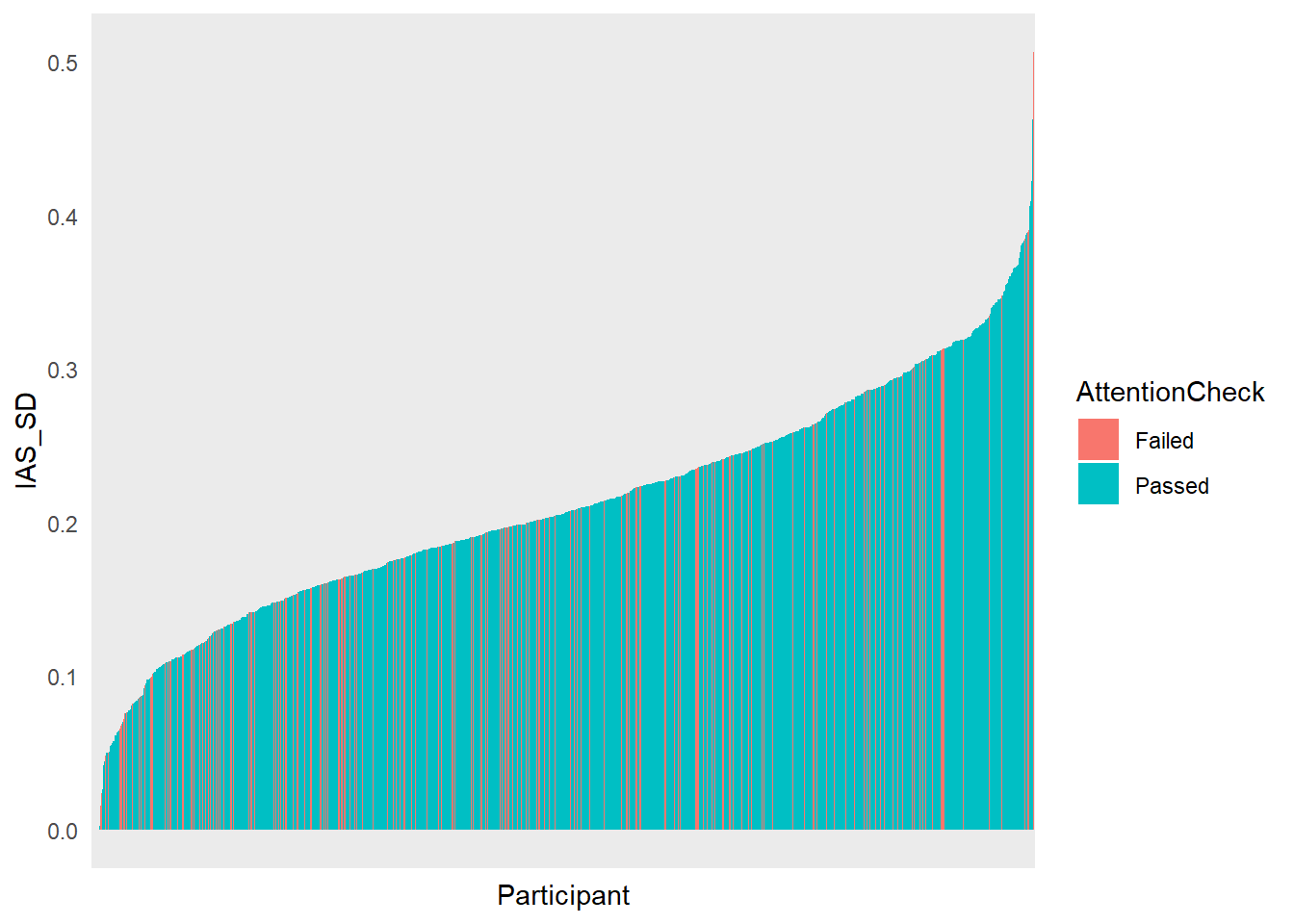
outliers$ias_sd <- as.character(df[abs(standardize(df$IAS_SD, robust=TRUE)) > qnorm(0.995), "Participant"])
outliers$ias_sd <- outliers$ias_sd[!outliers$ias_sd %in% c(outliers$attentionchecks, outliers$duration)]We removed 12 (1.14%) participants for failing being an outlier on the IAS’ SD value.
dfoutlier <- df |>
select(contains("IAS_"), -IAS_Duration, -IAS_Order, -IAS_Total, -IAS_SD) |>
# select(contains("BDI2_"), -BDI2_Duration, -BDI2_Order, -BDI2_Total, -BDI2_SD) |>
# select(contains("STAI5_"), -STAI5_Duration, -STAI5_Order, -STAI5_General, -STAI5_SD) |>
# select(contains("MAIA2_"), -MAIA_Duration, -MAIA_Order) |>
# datawizard::remove_empty_rows() |>
performance::check_outliers(
method=c("optics", "mahalanobis_robust"),
threshold=list(optics=2, optics_xi=0.1)) |>
as.data.frame() |>
mutate(Participant = fct_reorder(df$Participant, Distance_OPTICS),
Outlier_AttentionCheck = ifelse(Participant %in% outliers$attentionchecks, 1, 0),
Outlier_Distance = ifelse(abs(standardize(Distance_OPTICS, robust=TRUE)) > qnorm(0.995), 1, 0),
Experiment_Duration = df$Experiment_Duration,
Outlier = ifelse(Outlier_AttentionCheck == 1, "Failed Attention Checks", "Passed"))
outliers$distance <- as.character(dfoutlier[dfoutlier$Outlier_Distance == 1, "Participant"])
outliers$distance <- outliers$distance[!outliers$distance %in% c(outliers$attentionchecks, outliers$duration, outliers$ias_sd)]
m <- mgcv::gam(Outlier_AttentionCheck ~ s(Distance_OPTICS), data = dfoutlier, family = "binomial")
parameters::parameters(m)# Fixed Effects
Parameter | Log-Odds | SE | 95% CI | z | df | p
--------------------------------------------------------------------------
(Intercept) | -1.50 | 0.08 | [-1.66, -1.34] | -18.67 | 1051.00 | < .001
# Smooth Terms
Parameter | z | df | p
---------------------------------------------------
Smooth term (Distance OPTICS) | 7.05 | 1.00 | 0.008estimate_relation(m, length=40) |>
ggplot(aes(x=Distance_OPTICS, y=Predicted)) +
geom_ribbon(aes(ymin=CI_low, ymax=CI_high), alpha=0.2) +
geom_line() +
ggside::geom_xsidedensity(data=dfoutlier, aes(fill=Outlier), alpha=0.3, color=NA)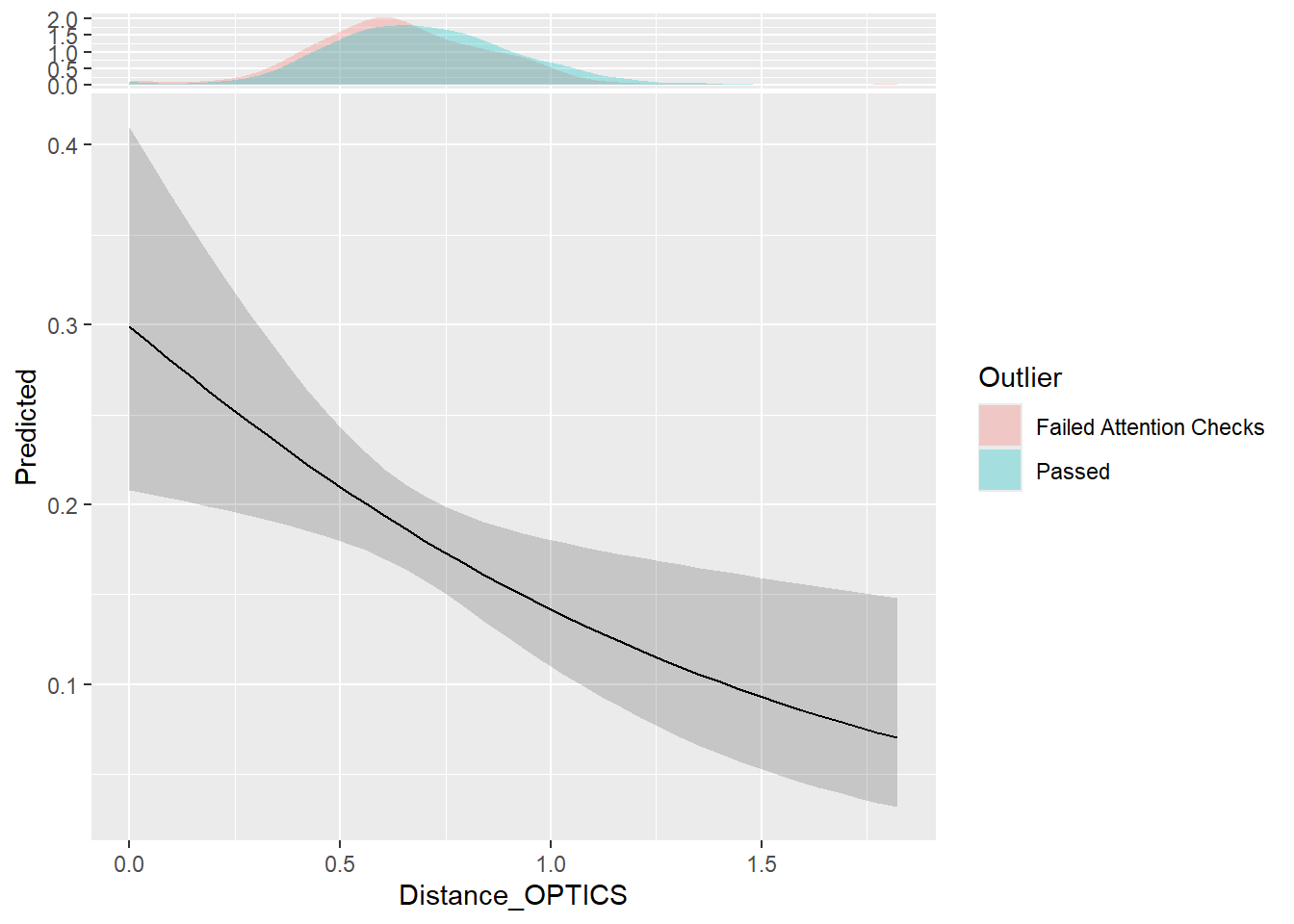
dfoutlier |>
ggplot(aes(x=Participant, y=Distance_OPTICS, fill=Outlier)) +
geom_bar(stat="identity") +
theme_minimal() +
theme(axis.text.x = element_blank())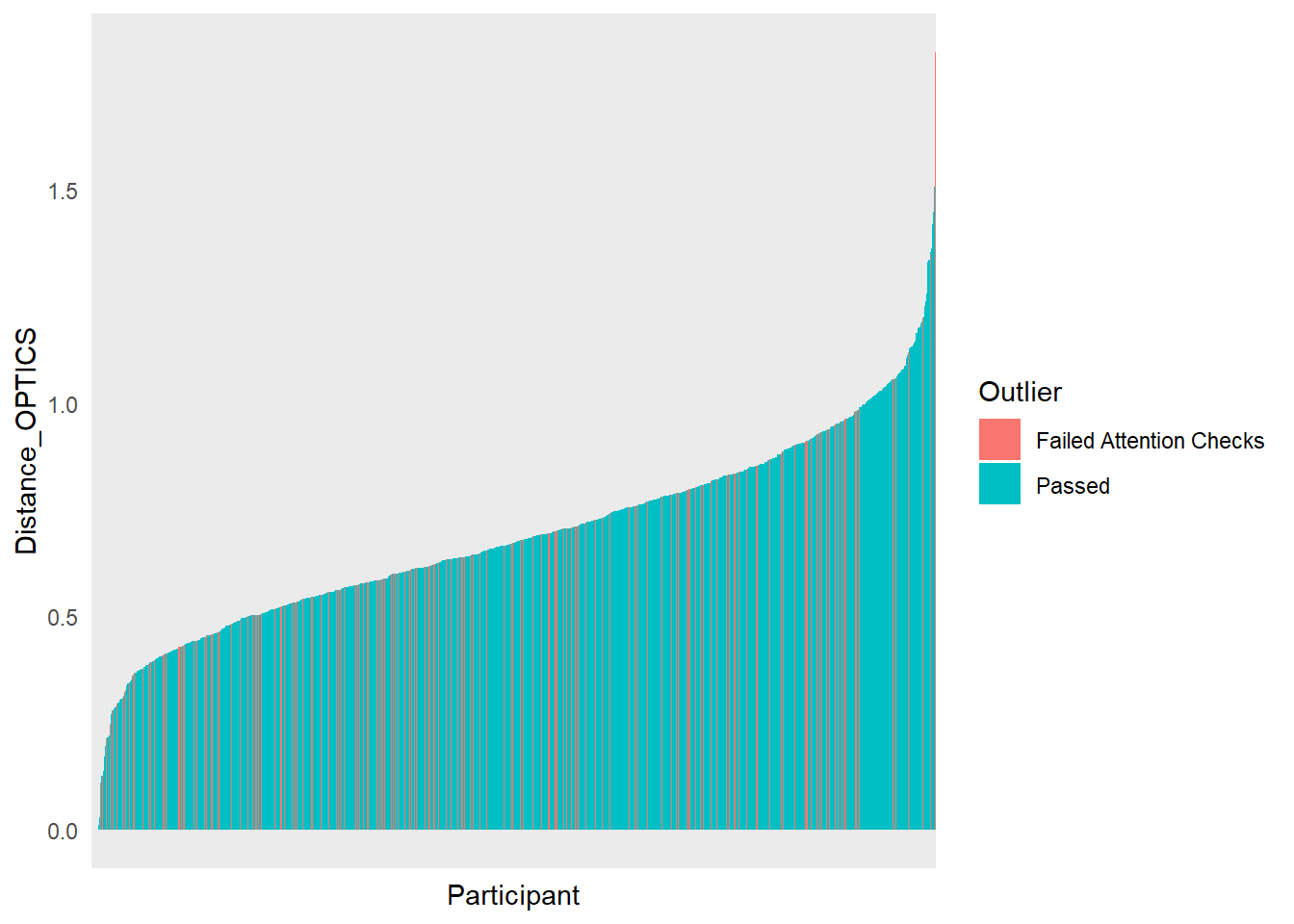
We removed 11 (1.04%) participants based on their multivariate distance. 23 (2.18%) outliers in total.
# length(c(outliers$ias_sd, outliers$distance))
# insight::format_percent(length(c(outliers$ias_sd, outliers$distance)) / nrow(df))
df <- filter(df, !Participant %in% c(outliers$attentionchecks, outliers$ias_sd, outliers$distance))
df <- select(df, -starts_with("AttentionCheck"))The final sample includes 836 participants (Mean age = 25.1, SD = 11.3, range: [18, 76], 0.1% missing; Gender: 73.8% women, 22.6% men, 2.87% non-binary, 0.72% missing; Education: Bachelor, 22.85%; Doctorate, 2.15%; High School, 63.52%; Master, 6.22%; missing, 0.24%; Other, 5.02%).
p_age <- df |>
mutate(Gender = ifelse(is.na(Gender), "Missing", Gender)) |>
ggplot(aes(x = x)) +
geom_histogram(aes(x = Age, fill=Gender), bins=28) +
geom_vline(xintercept = mean(df$Age), color = "red", linewidth=1.5) +
# geom_label(data = data.frame(x = mean(df$Age) * 1.15, y = 0.95 * 75), aes(y = y), color = "red", label = paste0("Mean = ", format_value(mean(df$Age)))) +
scale_fill_manual(values = c("Male"= "#64B5F6", "Female"= "#F06292", "Other"="orange", "Missing"="brown")) +
scale_x_continuous(expand = c(0, 0)) +
scale_y_continuous(expand = c(0, 0)) +
labs(title = "Age", y = "Number of Participants", color = NULL, subtitle = "Distribution of participants' age") +
theme_modern(axis.title.space = 10) +
theme(
plot.title = element_text(size = rel(1.2), face = "bold", hjust = 0),
plot.subtitle = element_text(size = rel(1.2), vjust = 7),
axis.text.y = element_text(size = rel(1.1)),
axis.text.x = element_text(size = rel(1.1)),
axis.title.x = element_blank()
)
p_ageWarning: Removed 1 row containing non-finite outside the scale range
(`stat_bin()`).Warning: Removed 1 row containing missing values or values outside the scale range
(`geom_vline()`).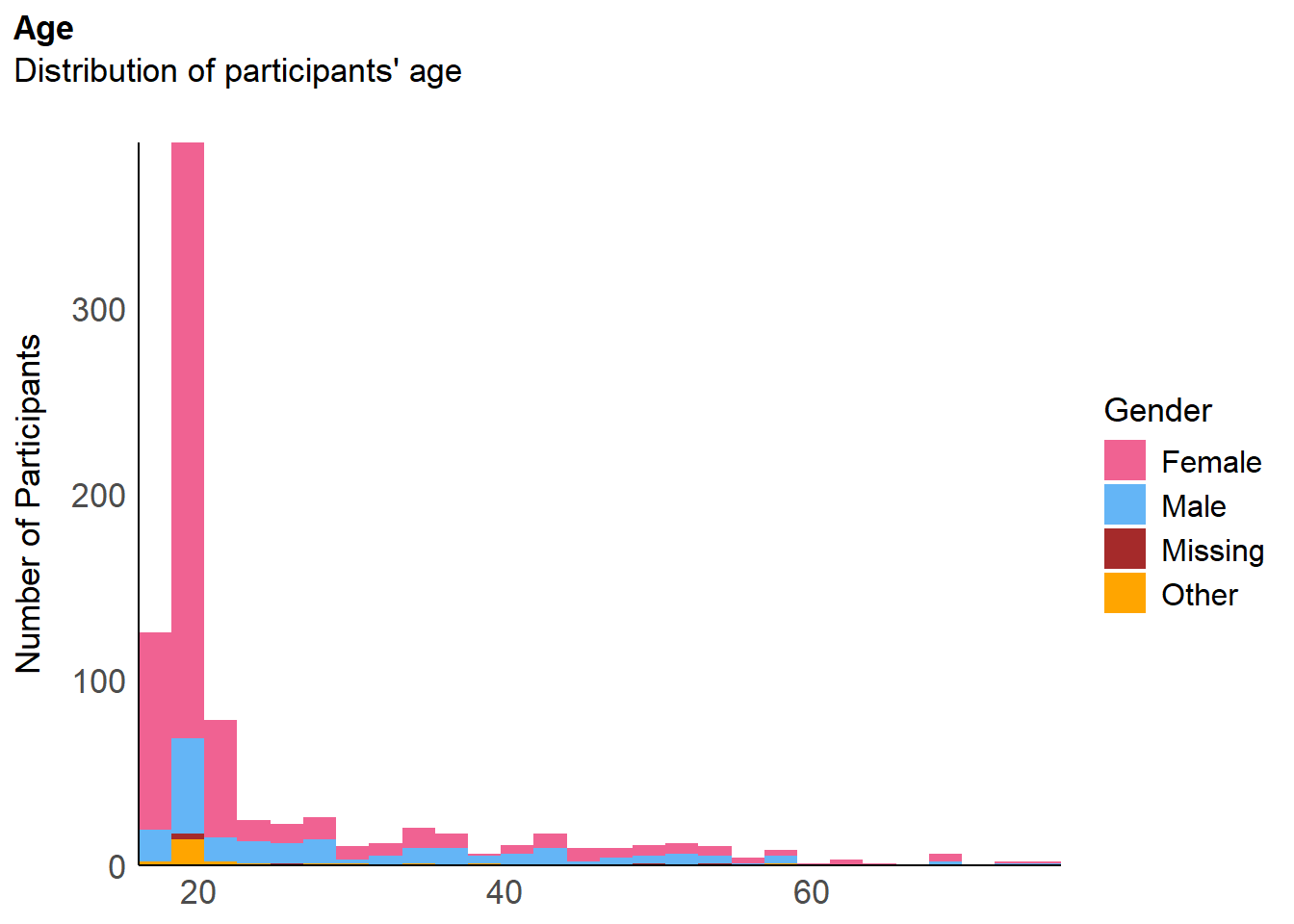
p_edu <- df |>
mutate(
Education = ifelse(Education == "", NA, Education),
Education = fct_relevel(Education, "Other", "High School", "Bachelor", "Master", "Doctorate")) |>
ggplot(aes(x = Education)) +
geom_bar(aes(fill = Education)) +
scale_y_continuous(expand = c(0, 0), breaks= scales::pretty_breaks()) +
scale_fill_viridis_d(guide = "none") +
labs(title = "Education", y = "Number of Participants", subtitle = "Participants per achieved education level") +
theme_modern(axis.title.space = 15) +
theme(
plot.title = element_text(size = rel(1.2), face = "bold", hjust = 0),
plot.subtitle = element_text(size = rel(1.2), vjust = 7),
axis.text.y = element_text(size = rel(1.1)),
axis.text.x = element_text(size = rel(1.1)),
axis.title.x = element_blank()
)
p_edu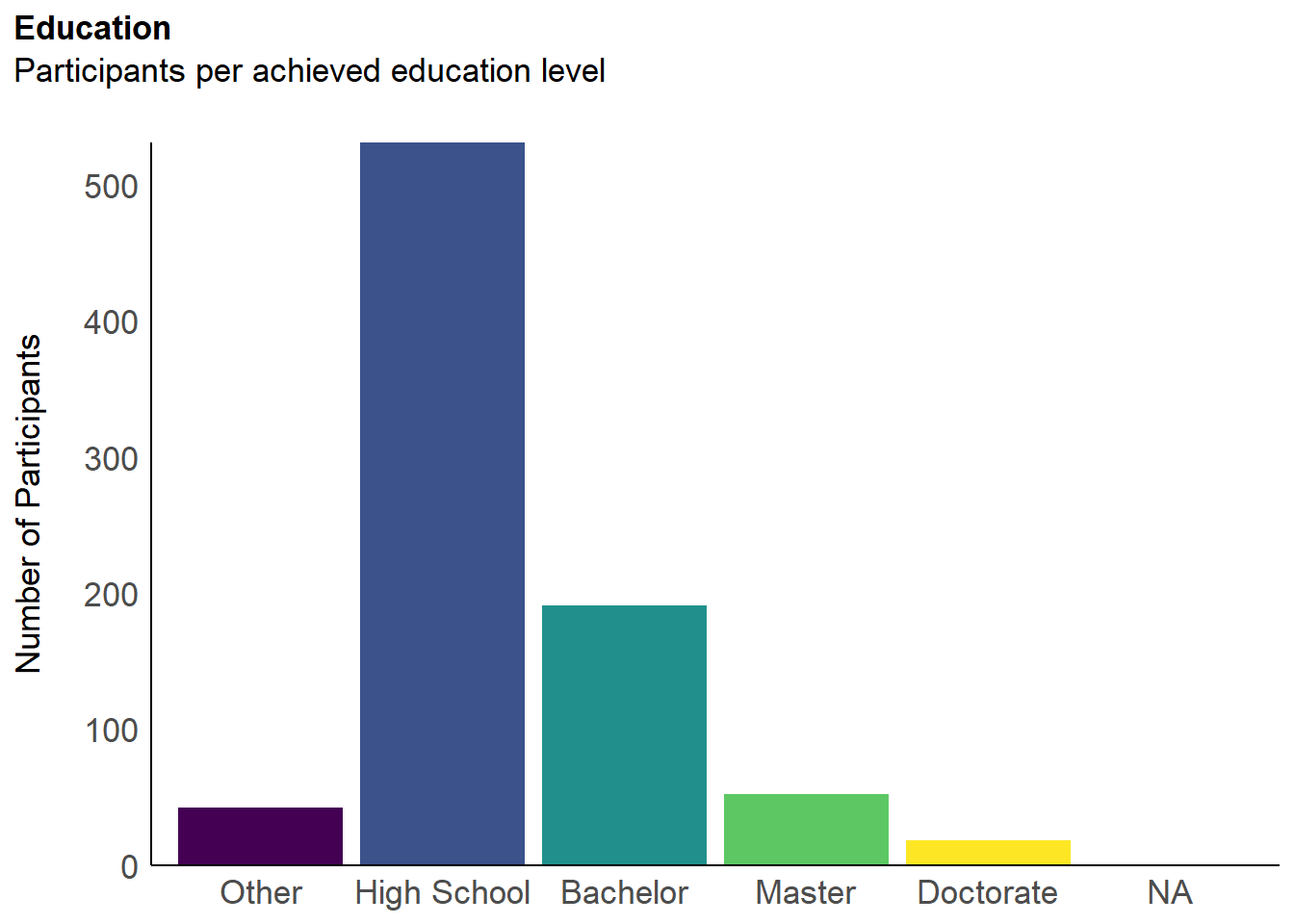
df$PHQ4_Duration[df$PHQ4_Duration > 3] <- NA
table1 <- df |>
select(Age,
PHQ4_Condition,
PHQ4_Total,
PHQ4_Duration,
STAI5_General,
BDI2_Total) |>
report::report_sample(by = "PHQ4_Condition")
table1$Variable <- str_remove_all(table1$Variable, fixed("Mean "))
table1$Variable <- str_remove_all(table1$Variable, fixed(" (SD)"))
table1$Variable <- c("Age", "PHQ4 (Total)", "PHQ4 (Duration)", "STAI5", "BDI2")add_bf <- function(df, var="PHQ4_Duration") {
dat <- df[, c(var, "PHQ4_Condition")]
dat <- dat[complete.cases(dat), ]
r <- report::report(t.test(dat[[var]] ~ dat[["PHQ4_Condition"]]), verbose=FALSE)
x <- as.data.frame(r)
f <- as.formula(paste(var, "~ PHQ4_Condition"))
x$BF <- as.data.frame(BayesFactor::ttestBF(formula=f, data=dat))$bf
x$Parameter <- var
x
}
table1_inference <- rbind(
add_bf(filter(df, !is.na(Age)), "Age"),
add_bf(df, var="PHQ4_Total"),
add_bf(df, var="PHQ4_Duration"),
add_bf(df, "STAI5_General"),
add_bf(df, "BDI2_Total")
) |>
select(Parameter, t, df_error, p, BF) |>
mutate(Parameter = str_remove_all(Parameter, fixed("df$")))
table1$BF <- table1_inference$BF
insight::display(table1)| Variable | PHQ4 - Original (n=396) | PHQ4 - Revised (n=440) | Total (n=836) | BF |
|---|---|---|---|---|
| Age | 25.48 (11.69) | 24.69 (10.91) | 25.07 (11.29) | 0.13 |
| PHQ4 (Total) | 4.70 (3.00) | 4.45 (3.00) | 4.57 (3.00) | 0.16 |
| PHQ4 (Duration) | 0.40 (0.30) | 0.40 (0.28) | 0.40 (0.29) | 0.08 |
| STAI5 | 1.57 (0.77) | 1.55 (0.77) | 1.56 (0.77) | 0.08 |
| BDI2 | 18.74 (11.84) | 17.63 (11.13) | 18.16 (11.47) | 0.20 |
df |>
select(PHQ4_Condition,
STAI5 = STAI5_General,
BDI2 = BDI2_Total,
PHQ4_Total) |>
standardize() |>
estimate_density(by = "PHQ4_Condition", method="KernSmooth") |>
ggplot(aes(x=x, y=y)) +
geom_line(aes(color = Parameter), linewidth=2) +
scale_color_manual(values = c("STAI5" = "#E91E63", "BDI2" = "#2196F3", "PHQ4_Total" = "#9C27B0")) +
labs(x = "Standardized Score", y = "Distribution") +
facet_wrap(~PHQ4_Condition) +
theme_bw()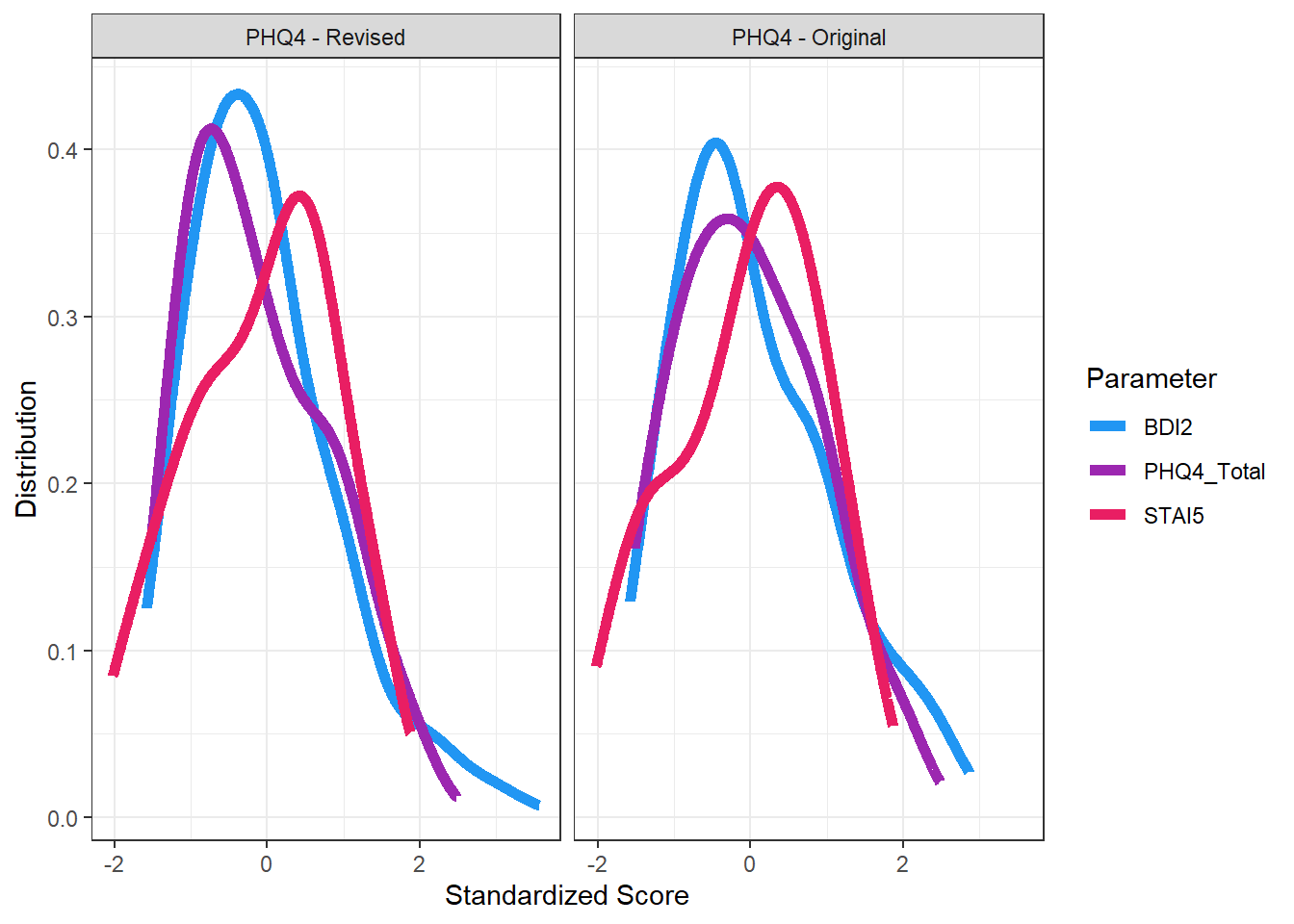
p1 <- select(df, Participant, Gender, starts_with("Disorder_")) |>
pivot_longer(cols = starts_with("Disorder_"), names_to = "Disorder", values_to = "Value") |>
mutate(Disorder = str_remove_all(Disorder, fixed("Disorder_"))) |>
summarize(N = sum(Value) / nrow(df), .by=c("Gender", "Disorder")) |>
mutate(N_tot = sum(N), .by="Disorder") |>
mutate(Disorder = fct_reorder(Disorder, desc(N_tot))) |>
ggplot(aes(x = Disorder, y = N, fill=Gender)) +
geom_bar(stat = "identity") +
scale_fill_manual(values = c("Male"= "#64B5F6", "Female"= "#F06292", "Other"="orange", "Missing"="brown")) +
scale_y_continuous(expand = c(0, 0), labels=scales::percent) +
labs(title = "Mood Disorders", y = "Number of Participants", subtitle = "Participants per self-declared mood disorder", x="Type") +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
theme_minimal()
p2 <- select(df, Participant, Gender, starts_with("DisorderTreatment_")) |>
pivot_longer(cols = starts_with("DisorderTreatment_"), names_to = "Treatment", values_to = "Value") |>
mutate(Treatment = str_remove_all(Treatment, fixed("DisorderTreatment_"))) |>
summarize(N = sum(Value) / nrow(df), .by=c("Gender", "Treatment")) |>
mutate(N_tot = sum(N), .by="Treatment") |>
mutate(Treatment = fct_reorder(Treatment, desc(N_tot))) |>
ggplot(aes(x = Treatment, y = N, fill=Gender)) +
geom_bar(stat = "identity") +
scale_fill_manual(values = c("Male"= "#64B5F6", "Female"= "#F06292", "Other"="orange", "Missing"="brown")) +
scale_y_continuous(expand = c(0, 0), labels=scales::percent) +
labs(title = "Treatment", y = "Number of Participants", subtitle = "Participants per treatment", x="Type") +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
theme_minimal()
p1 / p2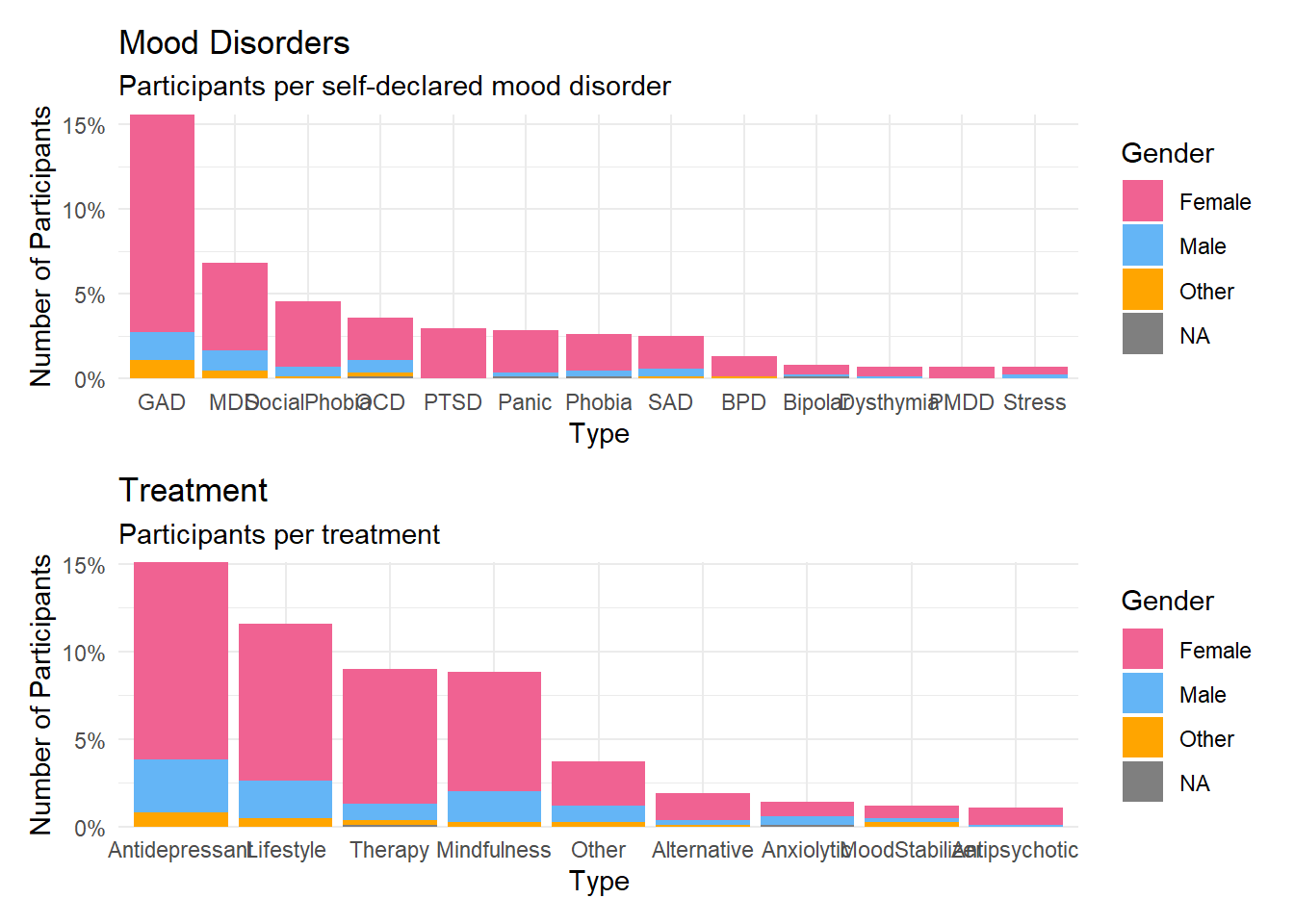
df$MoodDisorder <- rowSums(select(df, DisorderTreatment_MoodStabilizer, DisorderTreatment_Antidepressant, DisorderTreatment_Anxiolytic))
df$MoodDisorder <- ifelse(df$MoodDisorder > 0, TRUE, FALSE)
df$Treatment <- rowSums(select(df, DisorderTreatment_MoodStabilizer, DisorderTreatment_Antidepressant, DisorderTreatment_Anxiolytic, DisorderTreatment_Therapy))
df$Treatment <- ifelse(df$Treatment > 0, TRUE, FALSE)
df$Depression <- rowSums(select(df, Disorder_MDD, Disorder_Dysthymia))
df$Depression <- ifelse(df$Depression > 0 & df$Treatment, TRUE, FALSE)
df$Anxiety <- rowSums(select(df, Disorder_GAD, Disorder_Panic))
df$Anxiety <- ifelse(df$Anxiety > 0 & df$Treatment, TRUE, FALSE)
df$DisorderHistory <- ifelse(df$DisorderHistory == "Yes", TRUE, FALSE)write.csv(df, "../data/data.csv", row.names = FALSE)