NeuroKit2 0.1.5 'Complexity' is out 🎉
NeuroKit2 0.1.5 is out with greater support for complexity analysis of neurophysiological signals
NeuroKit2 0.1.5 is out! 🎉
In the latest 0.1.5 release of NeuroKit2, our team has fixed several bugs in existing functionalities and in particular, overhauled the support for computing complexity measures of neurophysiological signals. We added a ton of new indices of entropy and fractal dimensions, including:
- Petrosian’s, Katz’s and Sevcik fractal dimension
- Differential, Permutation, Spectral, SVD entropy
- Fisher information
- Hjorth’s and Lempel-Ziv’s complexity
- Relative Roughness
- Hurst and Lyapunov exponent(s)
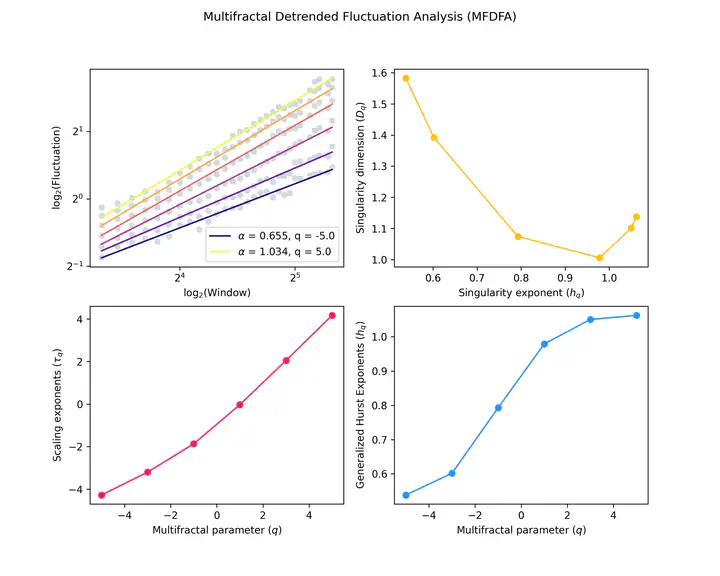
- Detrended Fluctuation Analysis (as well as MFDFA)
- …
You can compute them all using the new nk.complexity() function!
import neurokit2 as nk
signal = nk.signal_simulate(frequency=[5, 6], noise=0.5)
results, info = nk.complexity(signal, which="fast")
results
DiffEn FI Hjorth KFD PEn ... PFD RR SFD
1.536573 0.01524 1.355543 4.720953 1.0 ... 1.017423 1.638357 1.691036
To understand more about complexity science, we recommend reading our preprint, which introduces the theoretical (and mathematical) meanings of complexity and reviews the existing studies of complexity analysis across multiple fields of psychology.
Of course, the NeuroKit2 Python package also includes tons of other useful features for physiological signal processing (see this quick example)!
Don’t forget to watch our repo to keep a look out for more complexity functionalities coming up! 👀